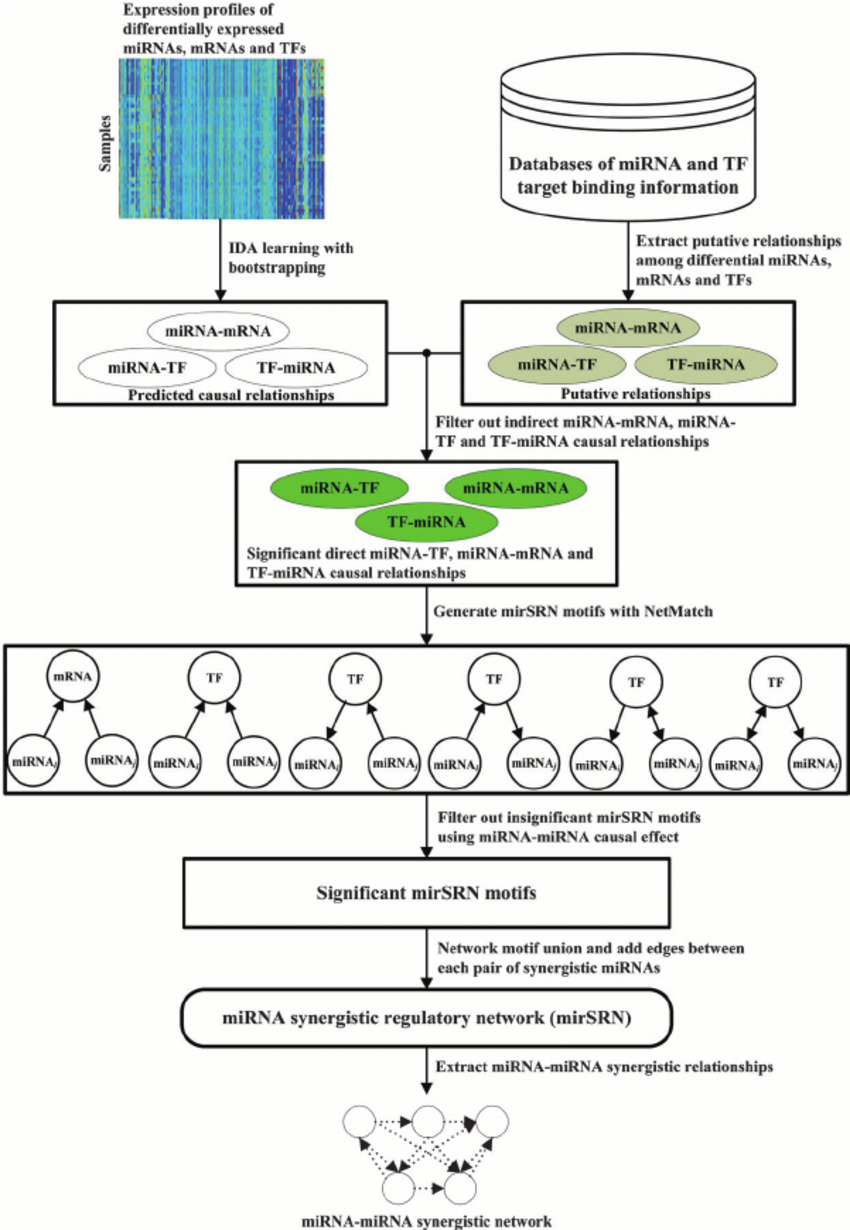
 A workflow of mirSRN
A workflow of mirSRN
 A workflow of mirSRN
A workflow of mirSRN
Understanding the synergism of multiple microRNAs (miRNAs) in gene regulation can provide important insights into the mechanisms of complex human diseases caused by miRNA regulation. Therefore, it is important to identify miRNA synergism and study miRNA characteristics in miRNA synergistic regulatory networks. A number of methods have been proposed to identify miRNA synergism. However, most of the methods only use downstream target genes of miRNAs to infer miRNA synergism when miRNAs can also be regulated by upstream transcription factors (TFs) at the transcriptional level. Additionally, most methods are based on statistical associations identified from data without considering the causal nature of gene regulation. In this paper, we present a causality based framework, called mirSRN (miRNA synergistic regulatory network), to infer miRNA synergism in human molecular systems by considering both downstream miRNA targets and upstream TF regulation. We apply the proposed framework to two real world datasets and discover that almost all the top 10 miRNAs with the largest node degree in the mirSRNs are associated with different human diseases, including cancer, and that the mirSRNs are approximately scale-free and small-world networks. We also find that most miRNAs in the networks are frequently synergistic with other miRNAs, and miRNAs related to the same disease are likely to be synergistic and in a cluster linked to a biological function. Synergistic miRNA pairs show higher co-expression level, and may have potential functional relationships indicating collaboration between the miRNAs. Functional validation of the identified synergistic miRNAs demonstrates that these miRNAs cause different kinds of diseases. These results deepen our understanding of the biological meaning of miRNA synergism.