 The workflow of miRLAB package
The workflow of miRLAB package
 The workflow of miRLAB package
The workflow of miRLAB package
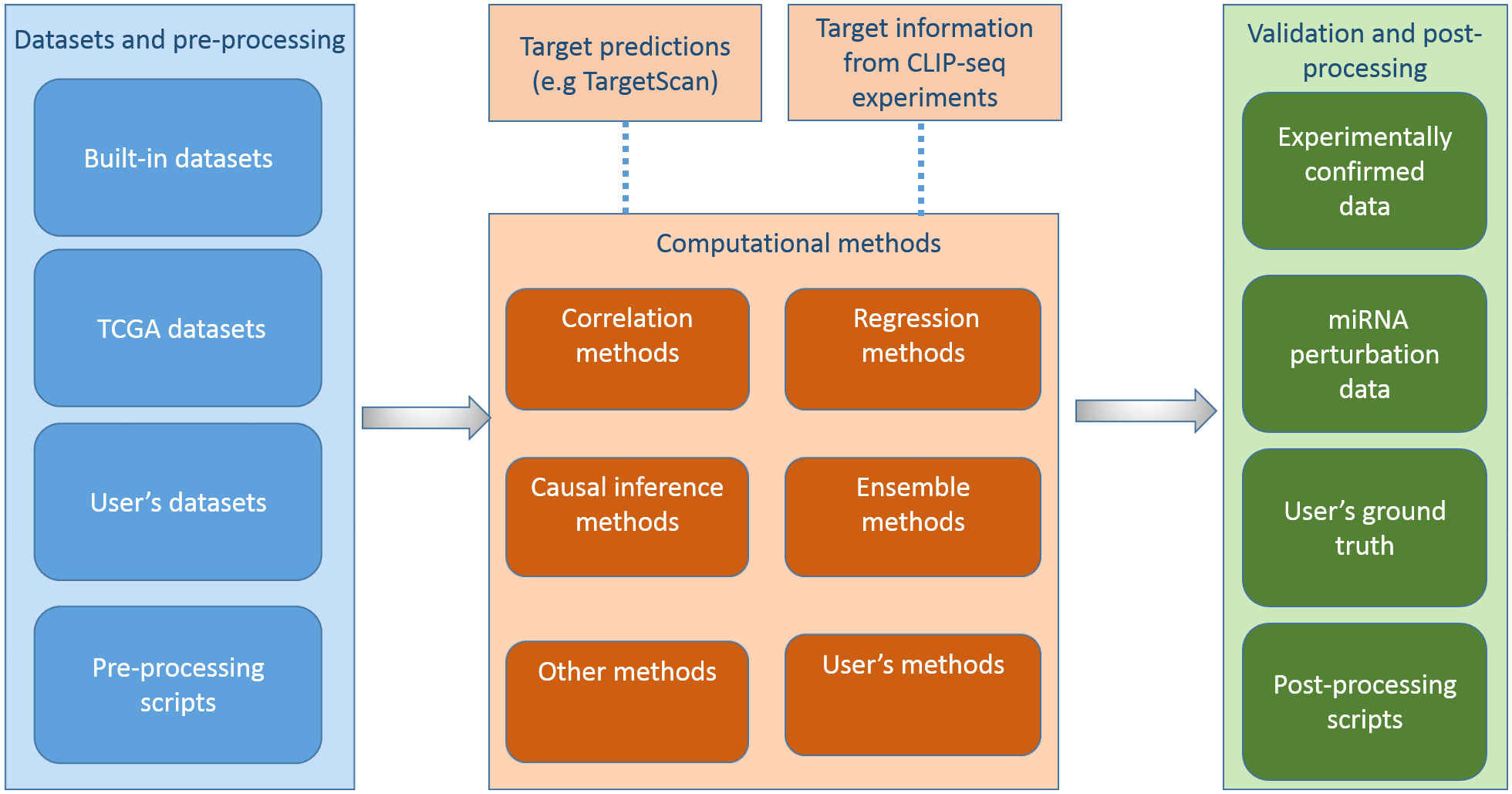
microRNAs (miRNAs) are important gene regulators at post-transcriptional level, and inferring miRNA-mRNA regulatory relationships is a crucial problem. Consequently, several computational methods of predicting miRNA targets have been proposed using expression data with or without sequence based miRNA target information. A typical procedure for applying and evaluating such a method is i) collecting matched miRNA and mRNA expression profiles in a specific condition, e.g. a cancer dataset from The Cancer Genome Atlas (TCGA), ii) applying the new computational method to the selected dataset, iii) validating the predictions against knowledge from literature and third-party databases, and comparing the performance of the method with some existing methods. This procedure is time consuming given the time elapsed when collecting and processing data, repeating the work from existing methods, searching for knowledge from literature and third-party databases to validate the results, and comparing the results from different methods. The time consuming procedure prevents researchers from quickly testing new computational models, analysing new datasets, and selecting suitable methods for assisting with the experiment design. Here, we present an R package, miRLAB, for automating the procedure of inferring and validating miRNA-mRNA regulatory relationships. The package provides a complete set of pipelines for testing new methods and analysing new datasets. miRLAB includes a pipeline to obtain matched miRNA and mRNA expression datasets directly from TCGA, 12 benchmark computational methods for inferring miRNA-mRNA regulatory relationships, the functions for validating the predictions using experimentally validated miRNA target data and miRNA perturbation data, and the tools for comparing the results from different computational methods.