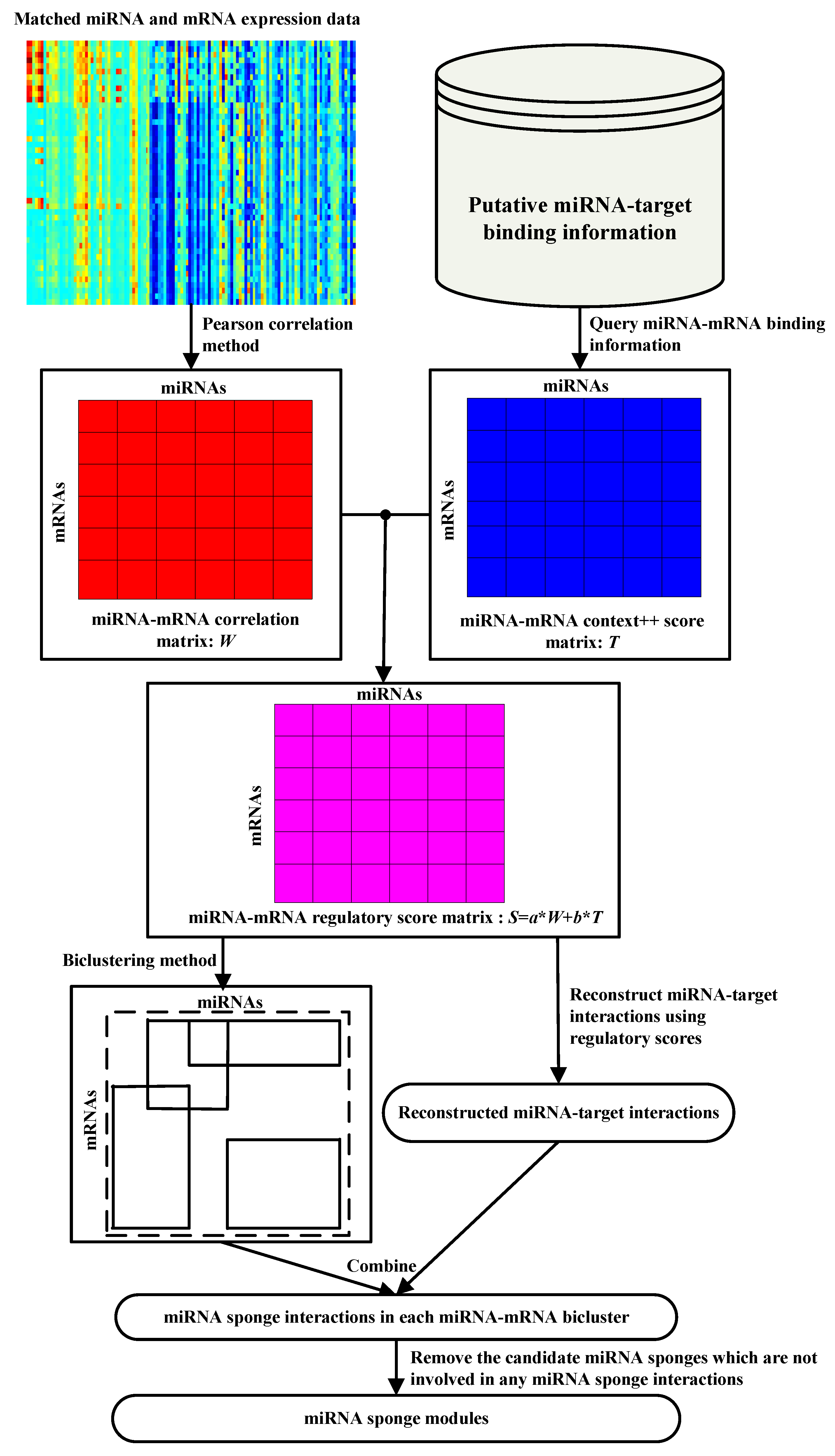
 The pipeline of miRSM
The pipeline of miRSM
 The pipeline of miRSM
The pipeline of miRSM
Background MicroRNA (miRNA) sponges with multiple tandem miRNA binding sequences can sequester miRNAs from their endogenous target mRNAs. Therefore, miRNA sponge acting as a decoy is extremely important for long-term loss-of-function studies both in vivo and in silico. Recently, a growing number of in silico methods have been used as an effective technique to generate hypotheses for in vivo methods for studying the biological functions and regulatory mechanisms of miRNA sponges. However, most existing in silico methods only focus on studying miRNA sponge interactions or networks in cancer, the module-level properties of miRNA sponges in cancer is still largely unknown. ResultsWe propose a novel in silico method, called miRSM (miRNA Sponge Module) to infer miRNA sponge modules in breast cancer. We apply miRSM to the breast invasive carcinoma (BRCA) dataset provided by The Cancer Genome Altas (TCGA), and make functional validation of the computational results. We discover that most miRNA sponge interactions are module-conserved across two modules, and a minority of miRNA sponge interactions are module-specific, existing only in a single module. Through functional annotation and differential expression analysis, we also find that the modules discovered using miRSM are functional miRNA sponge modules associated with BRCA. Moreover, the module-specific miRNA sponge interactions among miRNA sponge modules may be involved in the progression and development of BRCA. Our experimental results show that miRSM is comparable to the benchmark methods in recovering experimentally confirmed miRNA sponge interactions, and miRSM outperforms the benchmark methods in identifying interactions that are related to breast cancer. Conclusions Altogether, the functional validation results demonstrate that miRSM is a promising method to identify miRNA sponge modules and interactions, and may provide new insights for understanding the roles of miRNA sponges in cancer progression and development.